Abstract
Each case of Acute Myeloid Leukemia (AML) represents a unique ecosystem. Despite recent advances in our understanding of the genetic landscape of AML, this information remains insufficient to accurately match patients with targeted therapies. While pediatric and adult AML share phenotypic similarities, pediatric AML represents a genetically distinct disease from adult AML, and will benefit from independent genomic studies and novel therapeutic strategies. Real-time ex-vivo functional screening can identify mechanisms underpinning drug response and diversity between tumors, aiding in patient stratification. We established an in vitro drug screening system that incorporates cytokine signaling to better model inflammation, stem cell function, and niche derived support of leukemic blasts. This approach provided insight into variability between patients who currently would be placed on the same therapeutic regimen.
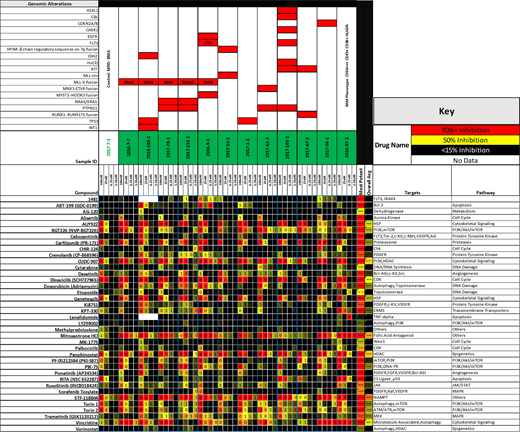
Samples were acquired from 12 pediatric AML patients, after informed consent was obtained. Samples were enriched for blasts, and cultured in the presence of SCF, TPO, FLT3-L, IL-3, and IL-6 (KTF36). A panel of 38 drugs was selected from a larger screen of 1839 compounds done on commercially available hematological malignancy cell lines. Drugs included standard chemotherapy agents used in AML and drugs currently under clinical development. Cells were exposed to drugs for 72hrs. An MTS assay was performed and results reported as % of viable cells remaining, after normalization to vehicle control wells. Targeted DNA NGS sequencing of 406 genes, 31 introns, and RNA sequencing of 265 genes was performed for genetic characterization.
In vitro drug screening revealed variations in drug sensitivity between samples and revealed time ex-vivo and cytokine milieu to be important factors affecting response of the same cells to the same drugs. Engraftment of long term cultures into immunodeficient mice produced aggressive disease in all cases, indicating robust support of stem cell function via addition of KTF36 cytokines. When possible, clinical response to therapy was compared with in vitro response to the same drugs. This screening approach highlighted well established agents that showed significant activity in highly refractory disease providing rationale for further clinical trial development. An unsupervised clustering showed drug sensitivity primarily correlated with the presence of MLL-X fusion, NRAS/KRAS and PTPN11 alterations. Linear Regression with interaction effects showed drug sensitivity/resistance to be highly selective for single/signature of specific molecular alterations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal